Description
Recognition site and hydrolysis position:
| DNA sequence with at least two 5mC: | DNA sequence with at least two 5mC: |
| RYN↑RY | RYN↑RY |
| YR↓NYR |
Source: Bacillus simplex 23
Substrate specificity:
Unit definition: One unit is defined as the amount of enzyme
required to hydrolyze at least one of three canonical sites
5`-G(5mC)NG(5mC)-3`/3`-CGN(5mC)G-5`
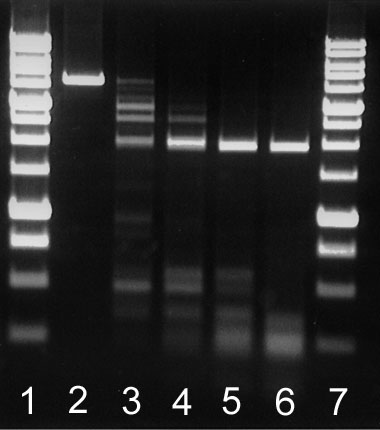
in 1 μg of linearized plasmid pFsp4HI3 in 1 hour at 30°C in a total reaction volume of 50 μl. As a result a linearized plasmid pFsp4HI3 disappears (see run 4 in the figure).
| BlsI activity assay on DNA pFsp4HI3/DriI Lanes: 1 and 7 – 1 Kb SE DNA-markers. 2 – Control DNA pFsp4HI3/DriI, 3 – 0.5 μl Bls I (1/5) 4 – 1 μl Bls I (1/5) 5 – 2 μl Bls I (1/5) 6 – 1 μl of undiluted BlsI Products were separated in 1,4% agarose gel in Buffer TAE. |
 |
Assayed on: DNA pFsp4HI3/DriI is a linearized plasmid pFsp4HI3, which carries a gene of DNA-methyltransferase M.Fsp4HI and includes three canonical sites:
5`-G(5mC)NG(5mC)-3`/3`-CGN(5mC)G-5` [2].
Optimal SE-buffer: SE-buffer W
Enzyme activity (%):
| B | G | O | W | Y | ROSE |
| 10 | 10 | 50 | 100 | 75 | 50 |
Storage conditions: 10 mM Tris-HCl (pH 7.5); 200 mM NaCl; 0.1 mM EDTA; 7 mM 2-mercaptoethanol; 200 μg /ml BSA; 50% glycerol; Store at -20°C.
Ligation: –
Non-specific hydrolisis: No detectable degradation of 1μg of Lambda DNA was observed after incubation with 5 units of enzyme for 16 hours at 30°C in a total reaction volume of 50 μl.
Methylation sensitivity: The enzyme cleaves C5-methylated DNA and doesn’t cut unmodified DNA [1].
Notes: References:
1. Chernukhin V.A., Tomilova J.E., Chmuzh E.V., Sokolova O.O., Dedkov V.S., Degtyarev S.Kh. Bacterial strain Bacillus simplex – producer of BlsI site specific endonuclease. // Russian Federation patent RU 2322494 C1 (2006).
2. Chmuzh E.V., Kashirina Yu.G., Tomilova Yu.E., Chernukhin V.A., Okhapkina S.S., Gonchar D. A., Dedkov V.S., Abdurashitov M. A., Degtyarev S. Kh. Gene cloning, comparative analysis of the protein structures from Fsp4HI restriction-modification system and biochemical characterization of the recombinant DNA methyltransferase M.Fsp4HI. // Molecular Biology, V.41, No 1, p. 43-50 (2007)
Application:
Degtyarev S.Kh., Prihod’ko G.G., Rechkunova N.I. Substrate specificity determination of restriction endonuclease SfeI.// Bioorg. chem (Moscow), Vol.14, No 6, 848-849 (1988). (In Russian)
Degtyarev S.Kh., Repin V.E., Rechkunova N.I. Proteus vulgaris – a strain producer of restriction endonuclease PvuII.// Soviet Union patent SU 1632974 (1991). (In Russian)
Kolykhalov A.A., Repin V.E., Fish A.M., Rechkunova N.I., Degtyarev S.Kh. Substrate specificity determination of restriction endonuclease Vha464I.// Sibirian Biological Journal, No 1, 60-61 (1991). (In Russian)
Degtyarev S. Kh., Kolyhalov A.A., Rechkunova N.I., Abdurashitov M.A. Acs I, a new restriction endonuclease from Arthrobacter citreus 310 recognizing 5'-Pu^AATTPy-3'. // Nucleic Acids Research, Vol. 20, No.14 (1992)
1. Chernukhin V.A., Tomilova J.E., Chmuzh E.V., Sokolova O.O., Dedkov V.S., Degtyarev S.Kh. Bacterial strain Bacillus simplex – producer of BlsI site specific endonuclease. // Russian Federation patent RU 2322494 C1 (2006).
2. Chmuzh E.V., Kashirina Yu.G., Tomilova Yu.E., Chernukhin V.A., Okhapkina S.S., Gonchar D. A., Dedkov V.S., Abdurashitov M. A., Degtyarev S. Kh. Gene cloning, comparative analysis of the protein structures from Fsp4HI restriction-modification system and biochemical characterization of the recombinant DNA methyltransferase M.Fsp4HI. // Molecular Biology, V.41, No 1, p. 43-50 (2007)
Application:
V.A. Chernukhin, Yu.E. Tomilova, E.V. Chmuzh, O.O. Sokolova, V.S. Dedkov, S.Kh. Degtyarev Site-specific endonuclease BlsI recognizes DNA sequence 5’-G(5mC)N^GC-3’ and cleaves it producing 3’ sticky ends // Translated from “Ovchinnikov bulletin of biotechnology and physical and chemical biology” V.3, No 1, pp 28-33, 2007
Chernukhin V.A, Abdurashitov M.A., Tomilov V.N., Gonchar D.A., Degtyarev S.Kh. Comparative analysis of mouse chromosomal DNA digestion with restriction endonucleases in vitro and in silico // Translated from “Ovchinnikov bulletin of biotechnology and physical and chemical biology” V.3, No 4, pp 19-27, 2007
D. A. Gonchar, A. G. Akishev, S. Kh. Degtyarev BlsI- and GlaI-PCR assays – a new method r
of DNA methylation studyr
// Translated from “Ovchinnikov bulletin of biotechnology and physical and chemical biology” V.6, No 1, pp 5-12, 2010
Alexander G. Akishev, Danila A. Gonchar, Murat A. Abdurashitov and Sergey Kh. Degtyarev Epigenetic typing of human cancer cell lines by BlsI- and GlaI-PCR assays // Translated from “Ovchinnikov bulletin of biotechnology and physical and chemical biology” V.7, No 3, pp 5-16, 2011



