Description
Recognition site and hydrolysis position:
| R(5mC)GY | R(5mC)↑GY |
| YG(5mC)R | YG↓(5mC)R |
Source: An E.coli strain that carries the cloned Gla I gene from Glacial ice bacterium Gl29
Substrate specificity:
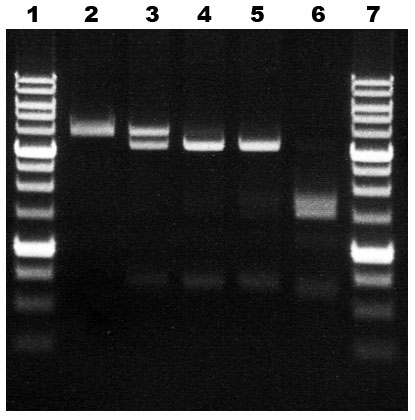
Unit definition: One unit is defined as the amount of enzyme required to hydrolyse completely a unique 5`-G(5mC)G(5mC)-3`/3`-(5mC)G(5mC)G-5` site in 1 μg of pHspAI2 plasmid DNA, which is linearized with GsaI, in 1 hour at 30°C in a total reaction volume of 50 μl. As a result of this site hydrolysis two main DNA fragments (3,419 and 699 bp) are produced (see lanes 3-5 in the figure). GlaI digestion of recognition sequences with three and two 5-methylcytosines results in additional bands appearance (lane 6 in the figure).
| GlaI activity assay on DNA pHspAI2/GsaI Lanes: 2 Control pHspAI2/GsaI DNA 3 – 0.5 μl GlaI (diluted 1/100), 4 – 1 μl GlaI (diluted 1/100), 5 – 2 μl GlaI (diluted 1/100), 6 – 1 μl of undiluted GlaI, 1 and 7- 1 Kb SE DNA Ladder.Products were separated in 1% agarose gel in TAE Buffer. |
 In the presence of 20% DMSO, the enzyme’s activity significantly increases without losing specificity In the presence of 20% DMSO, the enzyme’s activity significantly increases without losing specificity |
Assayed on: DNA pHspAI2/GsaI is a linearized plasmid pHspAI2, which carries a gene of DNA-methyltransferase M.HspAI (recognition sequence 5`-GCGC-3`) and includes a unique GlaI recognition site 5`-G(5mC)G(5mC)-3`/3`-(5mC)G(5mC)G-5` [2].
Optimal SE-buffer: SE-buffer Y
Enzyme activity (%):
| B | G | O | W | Y | ROSE |
| 75 | 75 | 25 | 25 | 100 | 100 |
Storage conditions: 10 mM Tris-HCl (pH 7.6); 250 mM NaCl; 0.1 mM EDTA; 7 mM 2-mercaptoethanol; 0,1% Triton X-100, 0.05 mg/ml BSA, 50% glycerol; Store at -20°C.
Ligation: –
Non-specific hydrolisis: No detectable degradation of 1μg of Lambda DNA was observed after incubation with 100 units of enzyme for 16 hours at 30°C in a total reaction volume of 50 μl.Reagents Supplied with Enzyme
10 X SE-buffer Y, pHspAI2/GsaI DNA
Methylation sensitivity: The enzyme cleaves only C5-methylated DNA and does not cut unmodified DNAand DNA with N4-methylcytosines [1].
Notes:
References:
- Chernukhin V.A., Nayakshina T.N., Tomilova J.E., Mezentseva N.V., Dedkov V.S., Degtyarev S.Kh. Bacterial strain Glacial ice bacterium I – producer of GlaI restriction endonuclease. // Russian Federation patent RU 2287012 C1 (2006).
- Valery A. Chernukhin, Tatyana N. Najakshina, Murat A. Abdurashitov, Julia E. Tomilova, Nina V. Mezentzeva, Vladimir S. Dedkov, Natalya A. Mikhnenkova, Danila A. Gonchar, Sergei Kh. Degtyarev A novel restriction endonuclease GlaI recognizes methylated sequence 5’-G(m5C)^GC-3’ // Biotechnologia (russ.). 2006. N 4. P. 31-35
- Degtyarev S.Kh., Belavin P.A., Repin V.E., Malygin E.G. Preparation method of restriction endonuclease Fok I from Flavobacterium okeanokoites // Soviet Union patent SU1436160 (1988). (In Russian)
- Degtyarev S.Kh., Rechkunova N.I., Grinev A.A., Dedkov V.S. Discovery and substrate specificity determination of restriction endonucleases Bme18I and Kzo9I.// Izvestia SB SA USSR, Biological sciences series, No.3, 25-26 (1989). (In Russian)
- Zemlyanskaya, E.V., Degtyarev, S.K. Substrate specificity and properties of methyl-directed site-specific DNA endonucleases.// Molecular Biology, v.47, No.6, p.900-913 (2013)
Application:
- , , , , , , , , , , , , , , , , , , , , Early Diagnosis of Colorectal Cancer Based on Bisulfite-free Site-specific Methylation Identification PCR Strategy: High-Sensitivity, Accuracy, and Primary Medical Accessibility. Adv. Sci. 2024, 2401137. https://doi.org/10.1002/advs.202401137
- Qiaomin Wu, Yang Yu, Mengqi Chen, Jinyan Long, Xiaolan Yang A label-free fluorescence sensing strategy based on GlaI-assisted EXPAR for rapid and accurate quantification of human methyltranferase activity // Talanta, 269, 125456 (2024)
- Xu, G., Yang, H., Qiu, J. et al. Sequence terminus dependent PCR for site-specific mutation and modification detection.// Nature Communications 14, 1169 (2023)
- Zhou S, Sun H, Huo D, Wang X, Qi N, Peng L, Yang M, Lu P, Hou C.A novel methyl-dependent DNA endonuclease GlaI coupling with double cascaded strand displacement amplification and CRISPR/Cas12a for ultra-sensitive detection of DNA methylation.// Analytica Chimica Acta, Volume 1212, 15 June 2022
- Hao Yang, Jiani Qiu, LinQing Zhen, Yizhou Huang, Wei Ren, Hongchen Gu, Hong Xu, Gaolian Xu.Sensitive GlaI digestion and terminal transferase PCR for DNA methylation detection// Talanta 2022 Sep 1;247:123616 (2022)
- Dong N, Wang W, Qin Y, Wang Y, Shan H.Sensitive lateral flow assay for bisulfite-free DNA methylation detection based on the restriction endonuclease GlaI and rolling circle amplification// Analytica Chimica Acta, Volume 1227, 22 September 2022
- Wang LJ, Han X, Qiu JG, Jiang B, Zhang CY.Cytosine-5 methylation-directed construction of a Au nanoparticle-based nanosensor for simultaneous detection of multiple DNA methyltransferases at the single-molecule level// Chem Sci. 2020, 11, 9675-9684 (2020)
- Petrova, D. V., Naumenko, M. B., Khantakova, D. V., Grin, I. R. & Zharkov, D. O. Relative Efficiency of Recognition of 5-Methylcytosine and 5-Hydroxymethylcytosine by Methyl-Dependent DNA Endonuclease GlaI// Russian Journal of Bioorganic Chemistry T. 45, № 6, стр. 625-629. (2019)
- Susan M Mitchell, Keith N Rand, Zheng-Zhou Xu, Thu Ho, Glenn S Brown, Jason P Ross, Peter L Molloy Helper-Dependent Chain Reaction (HDCR) for Selective Amplification of Methylated DNA Sequences // Methods Mol Biol (2018)
- Yueying Sun, Yuanyuan Sun, Weimin Tian, Chenghui Liu, * Kejian Gao and Zhengping L A novel restriction endonuclease GlaI for rapid and highly sensitive detection of DNA methylation coupled with isothermal exponential amplification reaction // Chemical Science, 9, pp.1344-1351 (2018)
- Viktor Tomilov, Murat Abdurashitov, Danila Gonchar, Anastasiya Snezhkina, George Krasnov, Anna Kudryavtseva and Sergey Degtyarev Comparative analysis of RCGY sites methylation in the genomes of Raji and U-937 malignant and normal lung fibroblast cell lines // Cancer Epigenetics and Biomarkers, Osaka, Japan, October 2017
- A.G. Akishev et al. GLAD-PCR assay of selected R(5mC)GY sites in URB1 and CEPBD genes in human genome// Res J Pharm Biol Chem Sci, 8(1): pp.465-475, (2017)
- Evdokimov et al. GLAD-PCR Assay of DNA Methylation Markers Associated with Colorectal Cancer// Biol Med (Aligarh) , 8:7(2016)
- A.A. Evdokimov, N.A. Netesova, N.A. Smetannikova, M.A. Abdurashitov, A.G. Akishev, E.S. Davidovich, Yu.D. Ermolaev, A.B. Karpov, A.E. Sazonov, R.M. Tahauov, S.Kh. Degtyarev Application of GLAD-PCR assay for determination of the methylation sites in the regulatory regions of tumor-supressors gene ELMO1 and ESR1 in colorectal cancer// Translated from Problems in oncology, #1, p.116-120 (2016).
- EV Dubinin, AG Akishev, MA Abdurashitov, SB Oleynikova, VL Sitko, and S Kh Degtyarev Real time GlaI-PCR assay of regulation regions of human genes HDAC4, RARB and URB1 // Research Journal of Pharmaceutical, Biological and Chemical Sciences, vol 7(2), p.p. 667-676 (2016).
- Murat A . Abdurashitov , Valery A . Chernukhin , Danila A . Gonchar , Vladimir S . Dedkov, Natalia A . Mikhnenkova, and Sergey Kh . Degtyarev Dimethyl Sulfoxide Changes the Recognition Site Preference of Methyl- directed Site-specific DNA Endonuclease GlaI// Research Journal of Pharmaceutical, Biological and Chemical Sciences, № 7(1),с. 1733-1739 (2016)
- Abdurashitov M.A., Tomilov V.N., Gonchar D.A., Kuznetsov V.V., Degtyarev S.Kh. Mapping of R(5mC)GY Sites in the Genome of Human Malignant Cell Line Raji // Biol Med (Aligarh) Volume 7, Issue 4, BM-135-15 (2015)
- Kuznetsov V.V., Abdurashitov M.A., Akishev A.G., Degtyarev S.H. Method for detection nucleotide sequence R(5mC)GY in given position of continuous DNA // Russian Federation patent RU 2525710 C1 (2014).
- Fagan RL, Wu M, Chedin F, Brenner C An Ultrasensitive High Throughput Screen for DNA Methyltransferase 1-Targeted Molecular Probes //PLoS ONE 8(11): e78752. doi:10.1371/journal.pone.0078752 (2013)
- Keith N. Rand, Graeme P. Young, Thu Ho and Peter L. Molloy, Sensitive and selective amplification of methylated DNA sequences using helper-dependent chain reaction in combination with a methylationdependent restriction enzymes. // Nucleic Acids Research, pp. 1-10 (2012).
- F. Syeda, R.L. Fagan, M. Wean, G.V. Awakumov, J.R. Walker, S. Xue, S. Dhe-Paganon, & C. Brenner, The RFTS Domain is a DNA-competitive Inhibitor of Dnmt1//, JBC, v. 286, pp. 15344-15351 (2011).
- Kravets AP, Mousseau TA, Litvinchuk AV, Ostermiller Sh, Vengen GS, Grodzinski DM. Changes in wheat DNA methylation pattern after chronic seed gamma-irradiation.// Tsitol Genet. 2010 Sep-Oct;44(5):18-22. Russian.
- Wood, R. J., McKelvie, J. C., Maynard-Smith, M. D., and Roach, P. L. A real-time assay for CpG-specific cytosine-C5 methyltransferase activity.// (2010) Nucleic Acids Res 38, e107
- Abdurashitov M.A., Chernukhin V.A, Gonchar D.A., Degtyarev S.Kh. GlaI digestion of mouse γ-satellite DNA: study of primary structure and ACGT sites methylation // BMC Genomics 2009, 10:322.



