Description
Recognition site and hydrolysis position:
| G(5mC)NG(5mC) | G(5mC)↑NG(5mC) |
| (5mC)GN(5mC)G | (5mC)GN↓(5mC)G |
Source: Glacial ice bacterium GL24
Substrate specificity:
Unit definition: One unit is defined as the amount of enzyme
required to hydrolyze completely a unique site:
5`-G(5mC)NG(5mC)-3`/3`-(5mC)GN(5mC)G-5`
in 1 μg of linearized plasmid pFsp4HI3/DriI in 1 hour at 37°C in a total reaction volume of 50 μl. As a result a linearized plasmid pFsp4HI3 disappears (see run 4 in the figure). GluI digestion of recognition sequences with three 5-methylcytosines results in additional bands appearance (runs 4-6 in the figure).
| GluI activity assay on DNA pFsp4HI3/DriI Lanes: 1 and 6 – 1 Kb SE DNA-markers. 2 – Control DNA pFsp4HI3/DriI, 3 – 0.5 μl Glu I 4 – 1 μl Glu I 5 – 2 μl Glu I Products were separated in 1,4% agarose gel in Buffer TAE. |
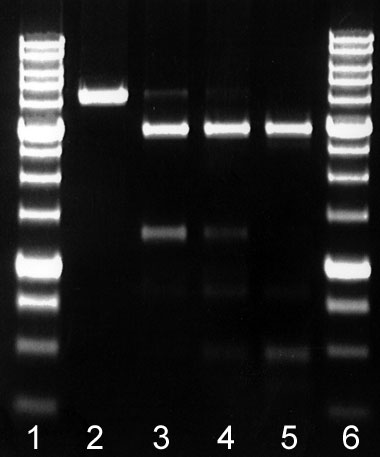 |
Assayed on: DNA pFsp4HI3/DriI is a linearized plasmid pFsp4HI3, which carries a gene of DNA-methyltransferase M.Fsp4HI and includes a unique GluI site:5`-G(5mC)NG(5mC)-3`/3`-(5mC)GN(5mC)G-5` [2].
Optimal SE-buffer: SE-buffer Y
Enzyme activity (%):
| B | G | O | W | Y | ROSE |
| 75 | 75 | 25 | 50 | 100 | 100 |
Storage conditions: 10 mM KH2PO4 (pH 7.45); 200 mM NaCl; 0.1 mM EDTA; 7 mM 2-mercaptoethanol; 200 μg/ml BSA, 50% glycerol; Store at -20°C.
Ligation: –
Non-specific hydrolisis: No detectable degradation of 1μg of Lambda DNA was observed after incubation with 1 units of enzyme for 16 hours at 37°C in a total reaction volume of 50 μl.
Methylation sensitivity: The enzyme cleaves C5-methylated DNAand does not cut unmodified DNA [1].
Notes: When using a buffer other than the optimal (suppied) SEBuffer, it may be necessary to add more enzyme to achieve complete digestion.
References:
1. Chernukhin V.A., Chmuzh E.V., Tomilova J.E., Nayakshina T.N., Dedkov V.S., Degtyarev S.Kh. Bacterial strain Glacial ice bacterium – producer of GluI site specific endonuclease. // Russian Federation patent RU 2322492 C1 (2006).
2. Zhilkina O.A., Repin V.E., Degtyarev S.Kh. Preparation method of restriction endonuclease SfaNI, that recognizes and cleaves DNA sequence 5′-GCATC(N)5^, 3′-CGTAG(N)9^.// Soviet Union patent SU 1442546 (1988). (In Russian)
Application:
Degtyarev S.Kh., Repin V.E., Rechkunova N.I. Proteus vulgaris – a strain producer of restriction endonuclease PvuII.// Soviet Union patent SU 1632974 (1991). (In Russian)
Kravets AP, Mousseau TA, Litvinchuk AV, Ostermiller Sh, Vengen GS, Grodzinski DM. Changes in wheat DNA methylation pattern after chronic seed gamma-irradiation.// Tsitol Genet. 2010 Sep-Oct;44(5):18-22. Russian.
1. Chernukhin V.A., Chmuzh E.V., Tomilova J.E., Nayakshina T.N., Dedkov V.S., Degtyarev S.Kh. Bacterial strain Glacial ice bacterium – producer of GluI site specific endonuclease. // Russian Federation patent RU 2322492 C1 (2006).
2. V.A. Chernukhin, E.V. Chmuzh, Yu.E. Tomilova, T.N. Nayakshina, D.A. Gonchar, V.S. Dedkov, S.Kh. Degtyarev A novel site-specific endonuclease GluI recognizes methylated DNA sequence 5’-G(5mC)^NG(5mC)-3’/3’-(5mC)GN^(5mC)G. // Translated from “Ovchinnikov bulletin of biotechnology and physical and chemical biology” V.3, No 2, pp 13-17, 2007
Application:
Chernukhin V.A, Abdurashitov M.A., Tomilov V.N., Gonchar D.A., Degtyarev S.Kh. Comparative analysis of mouse chromosomal DNA digestion with restriction endonucleases in vitro and in silico // Translated from “Ovchinnikov bulletin of biotechnology and physical and chemical biology” V.3, No 4, pp 19-27, 2007
Kravets AP, Mousseau TA, Litvinchuk AV, Ostermiller Sh, Vengen GS, Grodzinski DM. Changes in wheat DNA methylation pattern after chronic seed gamma-irradiation.// Tsitol Genet. 2010 Sep-Oct;44(5):18-22. Russian.