Description
Recognition site and hydrolysis position:
| (5mC)GNNNNNNN(5mC)G | (5mC)GNNNNN↑NN(5mC)G |
| G(5mC)NNNNNNNG(5mC) | G(5mC)NN↓NNNNNG(5mC) |
Source: An E.coli strain that carries the cloned Pcs I gene from Paracoccus carotinifaciens 3K.
Substrate specificity:
Unit definition:
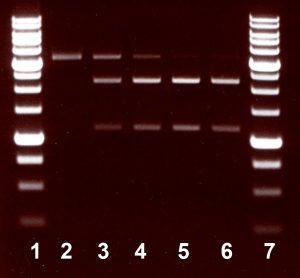
One unit is defined as the amount of enzyme required to digest 1 µg of pMHpySE49/DriI DNA in 1 hour at 37°C in a total reaction volume of 50 µl. As a result, a linearized plasmid pMHpySE49/DriI disappears, and two DNA fragments are produced.
Assayed on:
pMHpySE49 plasmid DNA, linearized with DriI (pMHpySE49/DriI).
pMHpySE49 (3536 bp) was obtained by a ligation of the vector pMTL22/FauNDI/SmaI and a PCR-fragment containing M.HpySE526I (forms 5′-A(5mC)GT-3′) gene and
DNA sequence:
5′-GACGTATAAAACGTT-3′
3′-CTGCATATTTTGCAA-5′
Thus, the plasmid has the optimal site of PcsI:
5′-A(5mC)GNNNNNNN(5mC)GT-3′
3′-TG(5mC)NNNNNNNG(5mC)A-5′
Optimal SE-buffer: SE-buffer 3M
Enzyme activity (%):
| B | G | O | W | Y | ROSE |
| 50-75 | 50-75 | 0 | 10-25 | 50-75 | 50 |
Storage conditions: 10 mM Tris-HCl (pH 7.5); 200 mM NaCl; 0.1 mM EDTA; 7 mM 2-mercaptoethanol; 0.1 mg/ml BSA, 50% glycerol; Store at -20°C.
Ligation: –
Non-specific hydrolisis: No detectable degradation of 1μg of Lambda DNA was observed after incubation with 1 units of enzyme for 16 hours at 37°C in a total reaction volume of 50 μl.
Reagents Supplied with Enzyme
10 X SE-buffer 3M
Methylation sensitivity: The enzyme cleaves only C5-methylated DNA and does not cut unmodified DNA [1].
Notes: When using a buffer other than the optimal (suppied) SEBuffer, it may be necessary to add more enzyme to achieve complete digestion.
References:
1. Chernukhin V.A., Nayakshina T.N., Tarasova M.V., Golikova L.N., Akishev A.G., Dedkov V.S., Mikhnenkova N.A., Degtyarev S.Kh. Bacterial strain Paracoccus carotinifaciens 3K- producer of PcsI site specific endonuclease. // Russian Federation patent RU 2377294 C1 (2009).